Installing and using iPAGE: a basic tutorial
Installation
Download the code (e.g, iPAGEvx.x.zip) to your machine. If you want, you can use wget to get the code:
wget https://tavazoielab.c2b2.columbia.edu/iPAGE/iPAGEvx.x.zip
Unzip the .zip file using unzip (files will be unzipped in a iPAGEvx.x/ directory)
unzip iPAGEvx.x.zip
Then, go to iPAGEvx.x/ and run make:
cd iPAGEvx.xmake
Finding informative pathways
The current implementation of iPAGE is meant to be executed from the same directory where all the scripts reside (in the iPAGEvx.x/ directory if you've followed the instructions above). If you want to run iPAGE from another directory, you should define PAGEDIR variable:
export PAGEDIR=/path/to/iPAGE/
If you don't set the PAGEDIR variable, iPAGE assumes the current path as the iPAGE home directory.
The basic command line syntax for iPAGE is :
perl page.pl --expfile=<inp> --species=<spp> --exptype=<typep>
where <inpp> indicates the input expression profile, <spp> indicates the species, and <typep> indicates whether the expression profile is discrete (e.g., cluster indices) or continuous (e.g., expression values obtained from a single microarray experiment).
For example, the following command line will run a test for an arbitrary continuous dataset :
perl page.pl --expfile=./TEST/bladder.exp --species=human --exptype=continuous --ebins=15
iPAGE creates an expfile_PAGE where the results are saved. The output files include:
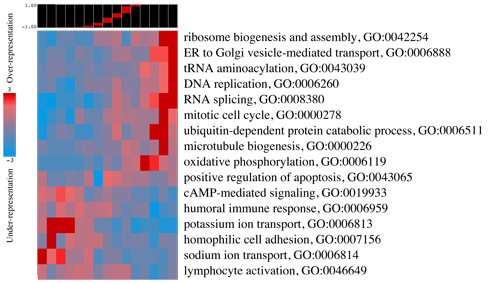
expfile.summary.pdf(eps) |
P-value heatmap combining significant categories (main figure) |
pvmatrix.txt |
Contains the data used to create the "expfile.summary.pdf" file |
pvmatrix.txt.killed |
Lists the categories that were significant but they were ommitted because of their redundancy |
Finally, here are some useful parameters that can be set for page.pl script:
--goindexfile=FILE --gonamesfile=FILE | Uses these files to run iPAGE instead of using "--species=sp" (defining --species would over-write these two parameters) |
--independence=1/0 | If set to 1, removes the redundant categories; otherwise, all the significant catgeories are included (default=1) |
--cattypes=F,P,C | For GO:terms you can change this parameter to "--cattypes=P,C" or any other combination |
--datafile=FILE --draw_sample_heatmap=true | In the "expfile.summary.pdf" it draws a heatmap of the average expression for each of the clusters. |
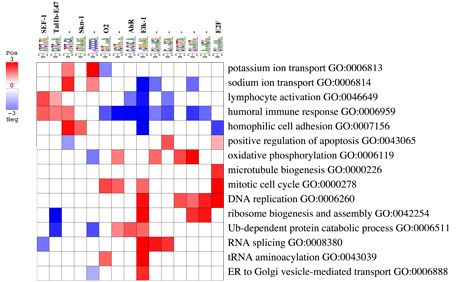
Buidling pathway motif interaction maps
The PRMG script (prmg.pl) is part of the iPAGE package and is located in the PAGEvx.x directory; however, it also relies on FIRE outputs to run (FIRE is available at https://tavazoielab.c2b2.columbia.edu/FIRE/):
export PAGEDIR=/path/to/iPAGE
perl prmg.pl --expfile=<inp> --species=<sp>
where <inp> indicates the input expression profile, <sp> indicates the species. The script does not work on expfile itself, but uses it to locate iPAGE and FIRE summary files (in expfile_PAGE and expfile_FIRE directories).
For example, the following command line will run PRMG on a continuous expression profile:
perl prmg.pl --expfile=./TEST/continuous.exp --species=human_go
The results are written to a motif_cat.cdt file and the graphical representations are created in the motif_cat.eps and motif_cat.pdf files.
For interactions with protein motifs (FIRE-pro results), prmg_protein.pl rather than prmg.pl should be used.